BIOC6240, GW
Next Generation Sequencing Technologies: Principles, Applications & Data Analysis
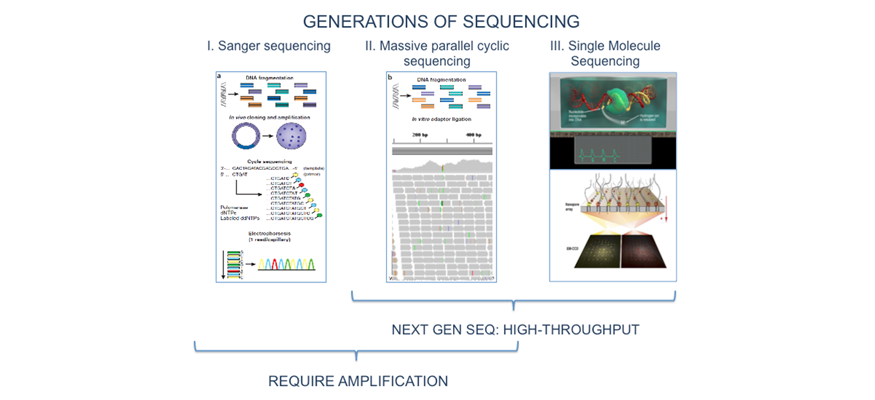
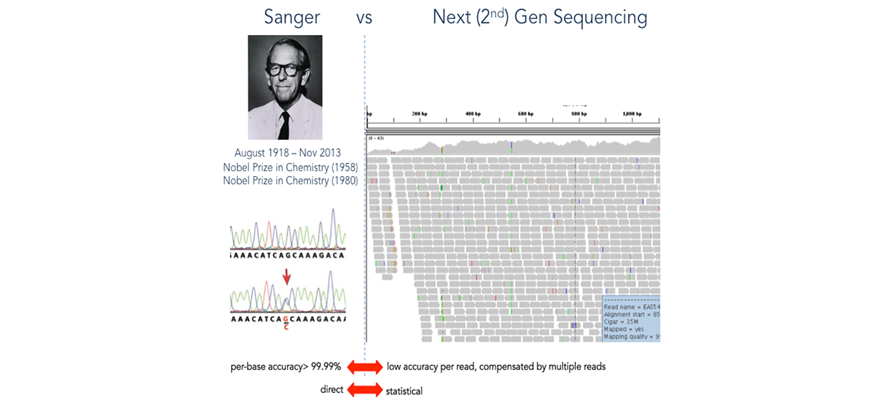
The demand for low-cost, high-throughput genome investigative studies has triggered the development of Next Generation Sequencing (NGS) technologies. Improved performance and decreasing costs have facilitated the development of a growing number of diverse NGS applications. The - $3-billion price tag for the first human genome in the 1990’s can now pay for a million human genome sequences, each completed within a few weeks. As a result, personal genome sequencing is becoming routine research and exome sequencing is being explored for diagnostic and therapeutic needs. NGS is expected to benefit many areas, ranging from molecular medicine, diagnostics, therapy design, and response prediction, to basic sciences such as evolutionary biology, energy production, agriculture, anthropology, and forensics.
To master this continuously expanding field requires novel educational approaches, and several efforts have been recently initiated by a number of leading University and Research centers. Next-Generation Sequencing Technologies: Principles, Applications, and Data Analysis has been developed to provide the graduate students theoretical basis and practical applications on NGS-based data generation, management, and analysis, as well as from A to Z experience with analyzing real human NGS datasets.
Justification: NGS applications are achieving a growing role in the diagnostics, prevention, and treatment of human diseases, requiring groundbreaking knowledge and expertise. This course is designed to provide students with theoretical and practical knowledge of NGS technologies and applications related to human health. The ultimate purpose of a comprehensive NGS educational program is a response to the field’s quickly expanding employment opportunities, for well-prepared scientists and physicians with vision on the NGS clinical and research applications.
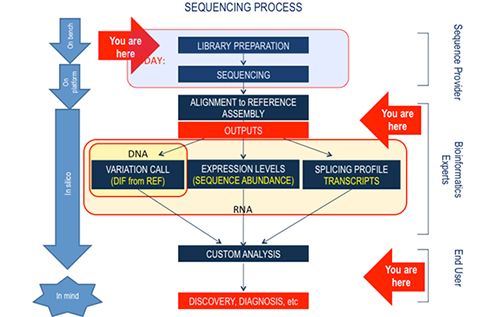
Description: “Next Generation Sequencing Technologies: Principles and Applications” consists of two major modules. The first module introduces the molecular principles underlying NGS technologies and provides the students with an understanding of possible research and clinical applications. It will also educate them about the major steps in the NGS process line - from the wet lab (library preparations), through “on instrument” processing (the actual sequencing process), to analysis and management of the raw and processed data.
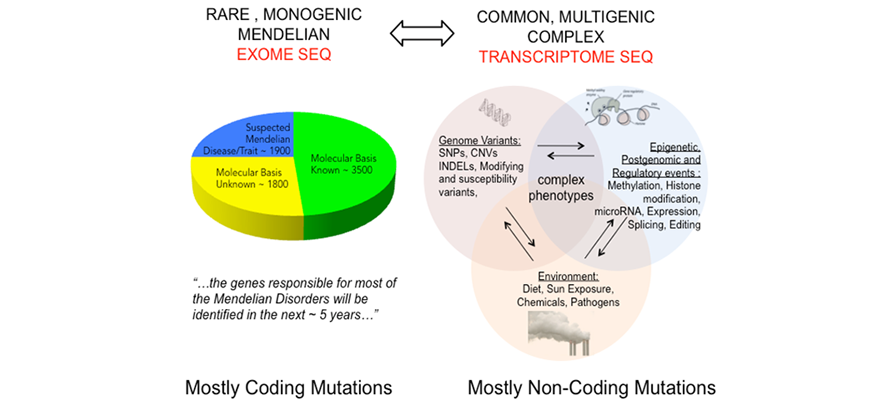
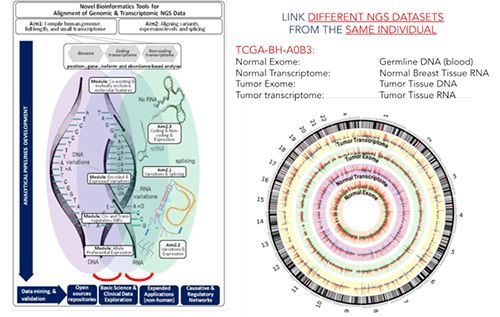
The second module is focused on the NGS analytical pipelines and data interpretation (includes labs and post-analytical data analysis and interpretation). It is designed to supply theoretical and practical knowledge about the analysis of NGS-generated datasets. Combined, the two modules will provide students with the ability to interpret results from NGS and to prioritize findings based on quality, statistical considerations, and biological meaningfulness. Students will be oriented into designing their own projects targeting specific scientific questions or clinical needs.
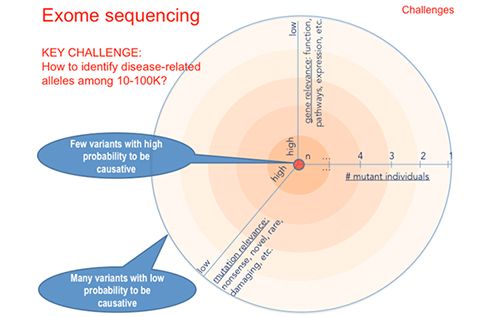
Objectives: By the end of the course, students are able to:
- Understand the molecular basis of NGS
- Have a detailed vision on NGS Applications and Opportunities in Science and Medicine
- Apply practical skills on NGS data analysis (from raw data to final output)
- Transform the outputs from the NGS platforms into biologically meaningful datasets
- Interpret NGS data
- Identify in silico genomics and bioinformatics’ tools for post-analytical data interpretation
- Extract, score, sort and filter NGS findings based on a) Technical quality b) Biological priority c) Potential Pathogenicity d) Statistical considerations
- Design NGS based approaches for clinical and research applications