Jump to: Ongoing Research Projects
Epigenetic modifications regulate gene expression and allow for tissue-specific expression of transcripts during development and in differentiated cells.
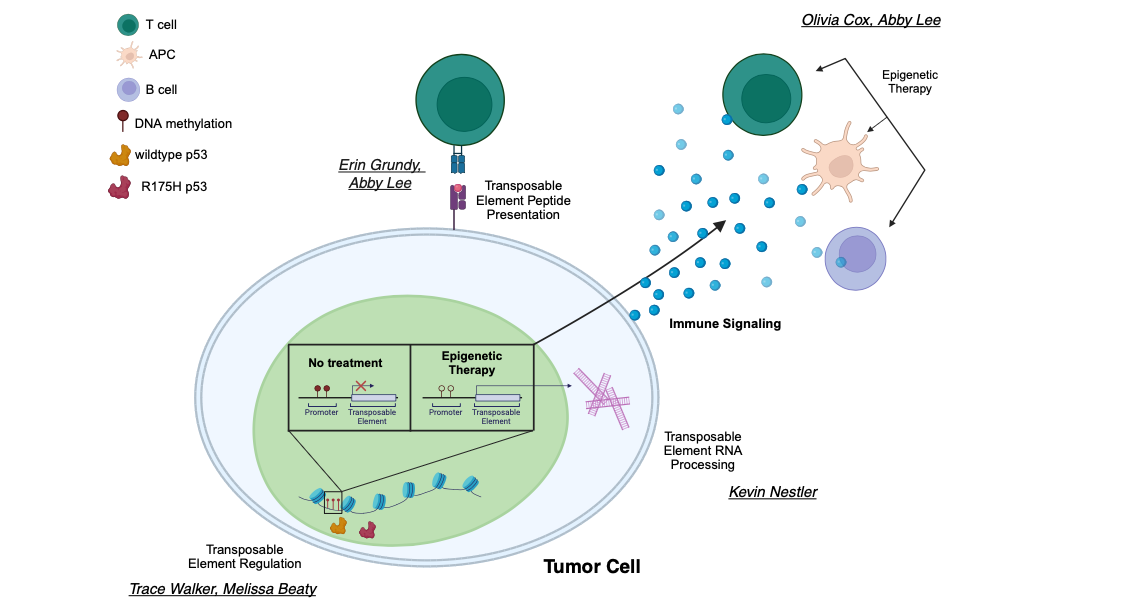
Cancer cells often have markedly different “epigenomes” than normal cells and exhibit profound changes in DNA methylation (a silencing epigenetic mark) of cytosines at CpG dinucleotides. These changes include global loss of methylation at regions such as repetitive elements that must be silenced for genome stability and gain of methylation at the promoter regions of tumor suppressor and other genes. DNA methyltransferase inhibitors (DNMTis) cause re-expression of genes that are silenced by promoter DNA methylation, reactivating tumor suppressor genes.
Transient exposure of multiple types of tumor cells to low doses of DNMTis promotes induction of apoptosis, reduced cell cycle activity, and decreased stem cell functions in cancer cells. Clinical efficacy of DNMTis such as 5-azacytidine (Aza) and 5-aza-2’-deoxycytidine has led to FDA approval for the pre-leukemic disorder myelodysplasia (MDS). Our work has shown that epigenetic therapy with DNMTis “boosts” immune signaling from tumors through activation of the interferon response by double-stranded RNA including hypermethylated endogenous retroviruses (ERVs). We have shown that a major effect of DNMTis is upregulation of immune signaling across solid tumors (breast, colon, and ovarian cancers), which holds true in patient samples from DNMTi clinical trials (Li, Chiappinell et al., Oncotarget 2014)
A key component of this dsRNA is endogenous retrovirus transcripts (ERVs), normally hypermethylated, which become demethylated and overexpressed upon DNMTi treatment. Working with Pamela Strissel and Reiner Strick at the University of Erlangen, we linked upregulation of hypermethylated ERVs to the DNMTi-induced immune response. Inhibiting the interferon response rescues about half of the apoptosis induced by 5-azacytidine, and that 5-azacytidine sensitizes a preclinical model of melanoma to anti-CTLA-4 (immune) therapy (Chiappinelli et al., Cell 2015). This epigenetic treatment upregulates interferon signaling in the tumor cells and causes host immune cells to selectively target the tumor cells for destruction.
Ongoing Research Projects
Our research focuses on epigenetic regulation of immune signaling in cancer. Epigenetic therapy is particularly appealing as it provides a way to alter gene expression without changing the DNA itself. Our lab studies the epigenetic changes in cancer and how epigenetic drugs can reverse these, specifically focusing on noncoding regions of the genome and the tumor cell immune response.
1. Which histone modifications regulate ERVs?
Our preliminary data shows synergistic activation of the interferon response in cancer cells when DNMTis are combined with inhibitors of other epigenetic modifiers. We are screening epigenetic drugs to determine which boost the interferon response most robustly. In parallel, we are performing chIP-Seq, genome-wide methylation analysis, and RNA-Seq on cancer cell lines with and without epigenetic drug treatments. These experiments allow us to test the hypothesis that both DNA methylation and histone modifications regulate ERV transcription, and, subsequently, the interferon response in cancer cells.
2. How does this affect activation of host immune cells?
DNMTis sensitize B16 melanoma tumors to anti-CTLA4 immunotherapy. Aza treatment of the MOSE ovarian cancer model increases recruitment of CD3+ T cells and CD8+IFNG+ T Effector cells to the tumor and combination treatment with Aza and anti-PD-1 reduces tumor burden. To test the hypothesis that interferon activation in cancer cells causes host immune cell recruitment and activation, we are manipulating cytosolic sensors, interferon pathway members, and cytokines to determine their role in 1) immune attraction and 2) sensitization to immune therapy in mouse models of tumorigenesis.
3. What is the role of dsRNA besides the ERVs in the Aza-induced interferon response?
Other repetitive RNAs (LINE, SINE elements) are upregulated by Aza and may contribute to the interferon response. We are combining strand-specific RNA-Seq results with RNA pulldowns with the MDA5 and TLR3 cytoplasmic sensors to determine which dsRNAs upregulated by Aza bind to these sensors. Specific dsRNA candidates or families will be overexpressed or knocked down to determine their functional role in the Aza interferon response. These experiments will test the hypothesis that other dsRNAs can be epigenetically upregulated and trigger the interferon response.